coming soon
• Umperovich, V., Béjà, O., Rozenberg*, A. The Practically ‘Complete’ Guide to Microbial Rhodopsins. in preparation
• Yaish, S., Pham, H.T., Kawasak, Y., Larom, S., Konno, M., Turner, M.S., Inoue, K., Béjà*, O., Rozenberg*, A. Suggested roles for heliorhodopsin in Lactococcus lactis strain 21. in preparation
• Nadel*, O., Hanna, R., Suissa-Szlejf, M., Sulimani, L., Keren, N., Adir, N., Béjà, O., Rozenberg*, A., Kleifeld*, O. An abundant uncultured marine cyanophage family encodes two distinct NblA proteins with differential activities toward different cyanobacterial phycobiliprotein subunits. in preparation
• Takaramoto, S., Zhao, C., Karasuyama, M., Schulz, F., Watanabe, M., Kawasaki, Y., Nagata, T., Konno, M., Morimoto, N., Fukuda, M., Inatsu, Y., Yawo, H., Béjà, O., Kato, H.K., Woyke*, T., Takeuchi*, I., Inoue*, K. Functional characterization of red-shifted rhodopsin channels from giant viruses explored by a machine-learning model for long-wavelength optogenetics. in preparation
see bioRxiv deposition
2026
• Marín*, M.d.C., Konno, M., Rozenberg, A., Béjà, O., Inoue*, K. Novel light-driven schizorhodopsins from Antarctic patescibacteria and cyanobacteria. Biophys. J. special issue: Retinal Proteins: Experiment and Computation. accepted
• Marín*, M.d.C., Murakoshi, S., Rozenberg, A., Tanaka, T., Konno, M., Shihoya*, W., Nureki*, O., Béjà*, O., Inoue*, K. (2026) Light-harvesting by antenna-containing xanthorhodopsin from an Antarctic Pseudanabaenaceae cyanobacterium. Commun. Biol. 9, 28.
2025

• Nadel, O., Hanna, R., Rozenberg, A., Shitrit, D., Tahan, R., Pekarsky, I., Béjà*, O., Kleifeld*, O., Lindell*, D. (2025) Viral NblA proteins negatively affect oceanic cyanobacterial photosynthesis. Nature 648, 434-442.
![]() see also our BEHIND THE PAPER story.
see also our BEHIND THE PAPER story.
• Galindo*, L.J., Takaramoto, S., Nagata, T., Rozenberg, A., Takahashi, H., Béjà, O., Inoue* K. (2025) Apusomonad rhodopsins, a new family of ultraviolet to blue light absorbing rhodopsin channels. Proc. Natl. Acad. Sci. USA 122, e2510619122.
• Mizutori, R., Katayama, K., Konno, M., Inoue, K., Béjà, O., Kandori*, H. (2025) Molecular properties of a viral heliorhodopsin, V2HeR2. Biophys. Physicobiol. 22, e220024.
• Das, I., Chazan, A., Church, J.R., Larom, S., León, R., Gómez-Villegas, P., Bárcenas-Pérez, D., Cheel, J., Koblížek, M., Béjà*, O., Schapiro*, I., Sheves*, M. (2025) Selective choice of the efficient carotenoid antenna by a xanthorhodopsin: controlling factors for binding and excitation energy transfer. JACS Au 5, 3070-3081.
• Takahashi, H., Takaramoto, S., Nagata, T., Fainsod, S., Kato, Y., Béjà, O., Inoue*, K. (2025) HulaChrimson: a new Chrimson-like cation channelrhodopsin discovered using freshwater metatranscriptomics from Lake Hula. Biophys. Physicobiol. 22, e220014.

• Tzlil, G., Marín, M.d.C., Matsuzaki, Y., Nag, P., Itakura, S., Mizuno, Y., Murakoshi, S., Tanaka, T., Larom, S., Konno, M., Abe-Yoshizumi, R., Molina-Márquez, A., Bárcenas-Pérez, D., Cheel, J., Koblížek, K., León, R., Katayama, K., Kandori, H., Schapiro, I., Shihoya*, W., Nureki*, O., Inoue*, K., Rozenberg*, A., Chazan*, A., Béjà*, O. (2025) Structural insights into light-harvesting by antenna-containing rhodopsins in marine Asgard archaea. Nat. Microbiol. 10, 1484-1500.
2024
• Takaramoto, S., Fainsod, S., Nagata, T., Rozenberg, A., Béjà, O., Inoue*, K. (2024) HulaCCR1, a pump-like cation channelrhodopsin discovered in a lake microbiome. J. Mol. Biol. 436, 168844.
• Wegner, H., Roitman, S., Kupczok, A., Braun, V., Woodhouse, J., Grossart, H-.P., Zehner, S., Béjà, O., Frankenberg-Dinkel*, N. (2024) Viral encoded Shemin pathway points towards the importance of tetrapyrrole metabolism during infection. Nat. Commun. 15, 8783.
• Mannen, K., Nagata, T., Rozenberg, A., Konno, M., Marín, M.d.C., Bagherzadeh, R., Béjà, O., Uchihashi, T., Inoue*, K. (2024) Multiple roles of a conserved glutamate residue for unique biophysical properties in a new group of microbial rhodopsins homologous to TAT rhodopsin. J. Mol. Biol. 436, 168331.
2023
• Oppermann, J., Rozenberg, A., Fabrin, T., Gonzalez Cabrera, C., Béjà, O., Prigge, M., Hegemann, P. (2023) Robust Optogenetic inhibition with red-light-sensitive anion-conducting channelrhodopsins. eLife 12, RP90100.
• N&V by Béjà, O., and Inoue, K. (2023) Iron-limitation light switch. Nat. Microbiol. 8, 1942-1943.
• Bar-Shalom, R., Rozenberg, A., Lahyani, M., Hassanzadeh, B., Sahoo, G., Haber, M., Burgsdorf, I., Tang, X., Squattrito, V., Gomez-Consarnau, L., Béjà, O., Steindler*, L. (2023) Rhodopsin-mediated nutrient uptake by cultivated photoheterotrophic Verrucomicrobiota. ISME J. 17, 1063-1073.

• Chazan, A., Das, I., Fujiwara, T., Shunya, M., Rozenberg, A., Molina-Márquez, A., Sano, F.K., Tanaka, T., Gómez-Villegas, P., Larom, S., Pushkarev, A., Malakar, P., Hasegawa, M., Tsukamoto, Y., Ishizuka, T., Konno, M., Nagata, T., Mizuno, Y., Katayama, K., Abe-Yoshizumi, R., Ruhman, S., Inoue, K., Kandori, H., León, R., Shihoya*, W., Yoshizawa*, S., Sheves*, M., Nureki*, O., Béjà*, O. (2023) Phototrophy by antenna-containing rhodopsin pumps in aquatic environments. Nature 615, 535-540.
 see also our BEHIND THE PAPER story.
see also our BEHIND THE PAPER story.

• Roitman*, S., Rozenberg, A., Lavy, T., Brussaard, C.P.D., Kleifeld, O., Béjà*, O. (2023) Isolation and infection cycle of a polinton-like virus virophage in an abundant marine alga. Nat. Microbiol. 8, 332-346.
 see also our BEHIND THE PAPER story.
see also our BEHIND THE PAPER story.
• Gyaltshen, Y., Rozenberg, A., Paasch, A., Burns, J.A., Warring, S., Larson, R.T., Maurer-Alcalá, X.X., Dacks, J., Narechania, A., Kim, E. (2023) Long-read–based genome assembly reveals numerous endogenous viral elements in the green algal bacterivore Cymbomonas tetramitiformis. Genome Biol. Evol. 15, evad194.
• Hanna, R., Rozenberg, A., Saied, L., Ben-Yosef, D., Lavy, T., Kleifeld O. (2023) In-depth characterization of apoptosis N-Terminome reveals a link between Caspase-3 cleavage and posttranslational N-Terminal acetylation. Mol. Cell. Proteomics 22, 100584.
• Hanna, R., Rozenberg, A., Lavy, T., Kleifeld, O. (2023) Increasing the coverage of the N-terminome with LysN amino terminal enrichment (LATE). Methods Enzymol. 686, 1-28.
• Salis, R.K., Schreiner, V.C., Rozenberg, A., Ohler, K., Baudy-Groh, P., Schäfer, R.B., Leese, F. (2023) Effects of fungicides on aquatic fungi and bacteria: a comparison of morphological and molecular approaches from a microcosm experiment. Environ. Sci. Eur. 35, 62.
2022
• Vierock*, J., Peter, E., Grimm, C., Rozenberg, A., Chen, I.W., Tillert, L., Castro Scalise, A.G., Casini, M., Augustin, S., Tanese, D., Forget, B.C., Peyronnet, R., Schnider-Warme, F., Emiliani, V., Béjà, O., Hegemann*, P. (2022) WiChR, a highly potassium selective channelrhodopsin for low-light one- and two-photon inhibition of excitable cells. Sci. Adv. 8, eadd7729.
• Hososhima, S., Mizutori, R., Abe-Yoshizumi, R., Rozenberg, A., Shigemura, S., Pushkarev, A., Konno, M., Katayama, K., Inoue, K., Tsunoda, S.P., Béjà, O., Kandori, H. (2022) Proton-transporting heliorhodopsins from marine giant viruses. eLife 11, e78416.
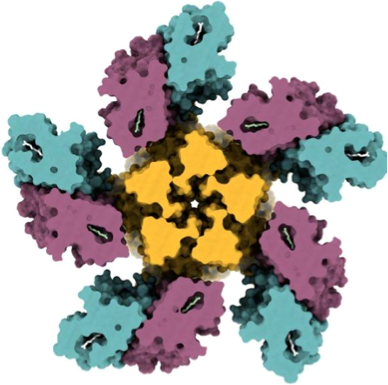
• Rozenberg, A., Kaczmarczyk, I., Matzov, D., Vierock, J., Nagata, T., Sugiura, M., Katayama, K., Kawasaki, Y., Konno, M., Nagasaki, Y., Toyama, M., Das, I., Pahima, E., Church, J., Adam, S., Borin, V.A., Chazan, A., Augustin, S., Wietek, J., Dine, J., Peleg, Y., Kawanabe, A., Fujiwara, Y., Yizhar, O., Sheves, M., Shapiro, I., Furutani, Y., Kandori, H., Inoue, K., Hegemann, P., Béjà*, O., and Shalev-Benami*, M. (2022) Rhodopsin-bestrophin fusion proteins from unicellular algae form gigantic pentameric ion channels. Nat. Struct. Mol. Biol. 29, 592-603.
• Rodriguez-Valera, F., Pushkarev, A., Rosselli, R., Béjà, O. (2022) Searching metagenoms for new rhodopsins. In: Gordeliy, V. (ed.) Rhodopsin. Methods Molecular Biology, 2501, 101-108.
• Jaffe, A.L., Konno, M., Kawasaki, Y., Kataoka, C., Béjà, O., Kandori, H., Inoue, K., Banfield, J.F. (2022) Saccharibacteria harness light energy using Type-1 rhodopsins that may rely on retinal sourced from microbial hosts. ISME J. 16, 2056-2059.
• Chazan, A., Rozenberg, A., Mannen, K., Nagata, T., Tahan, R., Yaish, S., Larom, S., Inoue, K., Béjà, O., Pushkarev*, A. (2022) Diverse heliorhodopsins detected via functional metagenomics in freshwater Actinobacteria, Chloroflexi and Archaea. Environ. Microbiol. 24, 110-121.
2021
• Rozenberg, A., Inoue, K., Kandori, H., Béjà, O. (2021) Microbial rhodopsins: the last two decades. Annu. Rev. Microbiol. 75, 427-447
• Dispatch by Arava, Y., Béjà, O. (2021) Phage biology: Stuck with dU. Curr. Biol. 31, R898-R899.
• Inoue*, K., Karasuyama, M., Nakamura, R., Konno, M., Yamada, D., Mannen, K., Nagata, T., Inatsu, Y., Yawo, H., Yura, K., Béjà, O., Kandori, H., Takeuchi*, I. (2021) Exploration of natural red-shifted rhodopsins by a machine-learning based Bayesian experimental design. Commun. Biol. 4, 362.
• Kataoka, C., Sugimoto, T., Shigemura, S., Katayama, K., Tsunoda, S.P., Inoue, K., Béjà, O., Kandori*, H. (2021) TAT rhodopsin is a UV-dependent environmental pH sensor. Biochemistry. 60, 899-907.
• Sahu, I., Mali, S. M., Sulkshane, P., Xu, C., Rozenberg, A., Morag, R., Sahoo, M., Singh, S.K., Ding, Z., Wang, Y., Day, S., Cong, Y., Kleifeld, O., Brik, A., Glickman, M.H. (2021) The 20S as a stand-alone proteasome in cells can degrade the ubiquitin tag. Nat. Commun. 12, 6173.
• Bock, N. A., Charvet, S., Burns, J., Gyaltshen, Y., Rozenberg, A., Duhamel, S., Kim, E. (2021) Experimental identification and in silico prediction of bacterivory in green algae. ISME J. 15, 1987-2000.
2020
• Hevroni*, G., Flores-Uribe, J., Béjà, O., Philosof*, A. (2020) Seasonal and diel patterns of abundance and activity of viruses in the Red Sea. Proc. Natl. Acad. Sci. USA. .
 see also our BEHIND THE PAPER story.
see also our BEHIND THE PAPER story.
• Rozenberg, A., Oppermann J., Wietek, J., Fernandez Lahore, R.G., Sandaa, R.-A., Bratbak, G., Hegemann*, P., Béjà*, O. (2020) Lateral gene transfer of anion-conducting channelrhodopsins between green algae and giant viruses. Curr. Biol. 30, 4910-4920.
 see also our BEHIND THE PAPER story.
see also our BEHIND THE PAPER story.
see Dispatch by Gallot-Lavallée, L., and Archibald, J.M. (2020) Evolution: Viral rhodopsins illuminate algal evolution.
• Inoue*, K., Tsunoda, S.P., Singh, M., Tomida, S., Hososhima, S., Konno, M., Nakamura, R., Watanabe, H., Uchihashi, T., Bulzu, P.-A., Banciu, H.L., Andrei, A.-Ş., Ghai, R., Béjà, O., Kandori*, H. (2020) Schizorhodopsins: A family of rhodopsins from Asgard archaea that function as light-driven inward H+ pumps. Sci. Adv. 6, eaaz2441.
• Pan, J., Zhou, Z., Béjà, O., Cai, M., Yang, Y., Liu, Y., Gu, J.-D., Li*, M. (2020) Genetic and transcriptomic evidence of light sensing, porphyrin biosynthesis, Calvin-Benson-Bassham cycle, and urea production in Bathyarchaeota. Microbiome 8, 43.
2019
• Nadel, O., Rozenberg, A., Flores-Uribe, J., Larom, S., Schwarz, R., Béjà*, O. (2019) An uncultured marine cyanophage encodes an active phycobilisome proteolysis adaptor protein NblA. Environ. Microbiol. Rep. 11, 848-854.
• Singh, M., Katayama, K. Béjà, O., Kandori*, H. (2019) Anion binding to mutants of the Schiff base counterion in heliorhodopsin 48C12. Phys. Chem. Chem. Phys. 21, 23663.

• Shihoya, W., Inoue, K., Singh, M., Konno, M., Hososhima, S., Yamashita, K., Ikeda, K., Higuchi, A., Izume, T., Okazaki, S., Hashimoto, M., Mizutori, R., Tomida, S., Yamaguchi, Y., Abe-Yoshizumi, R., Katayama, K., Tsunoda, S.P., Shibata, M., Furutani, Y., Pushkarev, A., Béjà, O., Uchihashi, T., Kandori*, H., Nureki*, O. (2019) Crystal structure of heliorhodopsin. Nature 574, 132-136.
• Karaoke, C., Inoue, K., Katayama, K., Béjà, O., Kandori*, H. (2019) Unique photochemistry observed in a new microbial rhodopsin. J. Phys. Chem. Lets. 10, 5117-5121.
• Oppermann, J., Liepe, B., Fischer, P., Rodriguez-Rozada, S., Flores-Uribe, J., Salipetre, A., , E., Keidel, A., Vierock, J., Kaufmann, J., Broser, M., Luck, M., Bartl, F., Hildebrandt, P., Wiegert, J.S., Béjà, O., Hegemann*, P., Wietek*, J. (2019) MerMAIDs: A novel family of metagenomically discovered, marine, anion-conducting and intensely desensitizing channelrhodopsins. Nature Commun. 10, 3315.
 see also our BEHIND THE PAPER story.
see also our BEHIND THE PAPER story.
• Yamaguchi, Y., Konno, M., Yamada, D., Yura, K., Inoue, K., Béjà, O., Kandori*, H. (2019) Engineered functional recovery of microbial rhodopsin without retinal-binding lysine. Photochem. Photobiol. 95, 116-1121.
• Flores-Uribe*, J., Philosof, A., Sharon, I., Fridman, S., Larom, S., Béjà*, O. (2019) A novel uncultured marine cyanophage lineage with lysogenic potential linked to a putative marine Synechococcus ‘relic’ prophage. Environ. Microbiol. Rep. 11, 598-604.
• Bulzu, P.-A., Andrei, A.-Ş., Salcher, M.M., Mehrshad, M., Inoue, K., Kandori, H., Béjà, O., Ghai*, R., Banciu, H.L. (2019) Casting light on Asgardarchaeota metabolism in a sunlit microoxic niche. Nat. Microbiol. 4, 1129-1137.
 see also our BEHIND THE PAPER story.
see also our BEHIND THE PAPER story.
• Tahara, S., Singh, M., Kuramochi, H., Shihoya, W., Inoue, K., Nureki, O., Béjà, O., Mizutani, Y., Kandori, H., Tahara*, T. (2019) Ultrafast dynamics of heliorhodopsins. J. Phys. Chem. B 123, 2507-2512.
• Flores-Uribe, J., Hevroni, G., Ghai, R., Pushkarev, A., Inoue, K., Kandori, H., Béjà*, O. (2019) Heliorhodopsins are absent in diderm (Gram-negative) bacteria: Some thoughts and possible implications for activity. Environ. Microbiol. Rep. 11, 419-424.
 see also our BEHIND THE PAPER story.
see also our BEHIND THE PAPER story.
• Dömel, J. S., Macher, T.-H., Dietz, L., Duncan, S., Mayer, C., Rozenberg, A., Wolcott, K., Leese, F., Melzer, R.R. (2019) Combining morphological and genomic evidence to resolve species diversity and study speciation processes of the Pallenopsis patagonica (Pycnogonida) species complex. Front. Zool. 16, 36.
2018

• Enav*, H., Kirzner, S., Lindell, D., Mandel-Gutfreund, Y., Béjà*, O. (2018) Adaptation to sub-optimal hosts is a driver of viral diversification in the ocean. Nature Commun. 9, 4698.
• Otomo, A., Mizuno, M., Singh, M., Shihoya, W., Inoue, K., Nureki, O., Béjà, O., Kandori, H., Mizutani*, Y. (2018) Resonance Raman investigation of the chromophore structure of heliorhodopsins. J. Phys. Chem. Lett. 8, 6431-6436.
• Singh, S., Inoue, K., Pushkarev, A., Béjà, O., Kandori*, H. (2018) Mutation study of heliorhodopsin 48C12. Biochemistry 57, 5041-5049.

• Pushkarev, A., Inoue, K., Larom, S., Flores-Uribe, J., Singh, M., Konno, M., Tomida, S., Ito, S., Nakamura, R., Tsunoda, S.P., Philosof, A., Sharon, I., Yutin, N., Koonin, E.V., Kandori*, K., Béjà*, O. (2018) A distinct abundant group of microbial rhodopsins discovered using functional metagenomics. Nature 558, 595-599.
 see also our BEHIND THE PAPER story.
see also our BEHIND THE PAPER story.
• Pushkarev, A., Hevroni, G., Roitman, S., Shim, J.-G., Choi, A., Jung, K.-H., Béjà*, O. (2018) The use of a chimeric rhodopsin vector for the detection of new proteorhodopsins based on color. Front. Microbiol. 9, 439.
• Ledermann, B., Schwan, M., Sommerkamp, J., Hofmann, E., Béjà, O., Frankenberg-Dinkel*, N. (2018) Evolution and molecular mechanisms of four-electron reducing ferredoxin-dependent bilin reductase from oceanic phages. FEBS J. 285, 339-356.
• Roitman, S., Hornung, E., Flores-Uribe, J., Sharon, I., Feussner, I., Béjà*, O. (2018) Cyanophage-encoded lipid-desaturases: oceanic distribution, diversity and function. ISME J. 12, 343-355.
2017

• Fridman, S., Flores-Uribe, J., Larom, S., Alalouf, O., Liran, O., Yacoby, I., Salama, F., Bailleul, B., Rappaport, F., Ziv, T., Sharon, I., Cornejo-Castillo, F.M., Philosof, A., Dupont, C.L., Sanchez, P., Acinas, S.G., Rohwer, F., Lindell*, D., Béjà*, O. (2017) A myovirus encoding both photosystem-I and II proteins enhances cyclic electron flow in infected Prochlorococcus cells. Nat. Microbiol. 2, 1350-1357.
 see also BEHIND THE PAPER.
see also BEHIND THE PAPER.
• Philosof*, A., Yutin, N., Flores-Uribe, J., Sharon, I., Koonin, E.V., Béjà*, O. (2017) Novel abundant oceanic viruses of uncultured Marine Group II Euryarchaeota. Curr. Biol. 27, 1362-1368.
• Yaniv, K., Golberg, K., Kramarsky-Winter, E., Marks, R., Pushkarev, A., Béjà, O., Kushmaro*, A. (2017) Functional marine metagenomic screening for anti-quorum sensing and anti-biofilm activity. Biofouling 33, 1-13.
• Macher, J. N., Leese, F., Weigand, A. M., Rozenberg*, A. (2017) The complete mitochondrial genome of a cryptic amphipod species from the Gammarus fossarum complex. Mitochondrial DNA Part B 2, 17-18.
2016
• Pinhassi, J., DeLong, E.F., Béjà, O., Gonzalez, J.M., Pedros-Alio, C. (2016) Marine bacterial and archaeal ion-pumping rhodopsins: genetic diversity, physiology and ecology. Microbiol. & Mol. Biol. Rev.80, 929-954.
• Ledermann, B., Béjà, O., Frankenberg-Dinkel, N. (2016) New biosynthetic pathway for pink pigments from uncultured oceanic viruses. Environ. Microbiol. 18, 4337-4347.
• Pushkarev, A., and Béjà, O. (2016) Functional metagenomic screen reveals new and diverse microbial rhodopsins. ISME J. 10, 2331-2335.
2015
• Roitman, S., Flores-Uribe, J., Philosof, A., Knowles, B., Rohwer, F., Ignacio-Espinoza, J.C., Sullivan, M.B., Cornejo-Castillo, F.M., Sanchez, P., Acinas, S.G., Dupont, C.L., Béjà*, O. (2015) Closing the gaps on the viral photosystem-I psaDCAB gene organization. Environ. Microbiol. 17, 5100-5108.
• Hevroni, G., Enav, H., Rohwer, F., Béjà*, O. (2015) Diversity of viral photosystem-I psaA genes. ISME J. 9, 1892-1898.
2014
• Beja O. and Lanyi, J.K. (2014) Nature’s toolkit for microbial rhodopsin ion pumps. Proc. Natl. Acad. Sci. USA 111, 6538-6539.
• Enav, H., Mandel-Gutfreund, Y., Béjà, O. (2014) Comparative metagenomic analyses reveal viral induced shifts of host metabolism towards nucleotide biosynthesis. Microbiome 2, 9.
2013
• Sabehi, G., and Béjà, O. (2013) Preparation of BAC libraries from marine microbial populations. Methods Enzymol. 531, 111-122.
• Philosof, A., and Béjà, O. (2013) Bacterial, archaeal and viral-like rhodopsins from the Red Sea. Environ. Microbiol. Rep. 5, 475-482.
• Béjà, O., Pinhassi, J., Spudich, J.L. Proteorhodopsins: widespread microbial light-driven proton pumps. In: Levin S.A. (ed.) Encyclopedia of Biodiversity 6, 280-285.
• Finkel, O.M., Béjà, O., Belkin, S. (2013) Global abundance of microbial rhodopsins. ISME J. 7, 448-451.
2012
• Mazor, Y., Greenberg, I., Toporik, H., Béjà, O., Nelson, N. (2012) The evolution of PSI in light of phage encoded reaction centers. Phil. Trans. R. Soc. B. 367, 3400-3405.
• Bodaker, I., Suzuki, M.T., Oren, O., Béjà, O. (2012) Dead Sea rhodopsins revisited. Environ. Microbiol. Rep. 4, 617-621.
• Béjà, O., Fridman, S., Glaser, F. (2012) Viral clones from the GOS expedition with an unusual photosystem-I gene cassette organization. ISME J. 6, 1617-1620.
• Atamna-Ismaeel, N., Finkel, O.M., Glaser, F., von Mering, C., Vorholt, J.A., Koblizek, M., Belkin, S., Béjà, O. (2012) Bacterial anoxygenic photosynthesis on plant leaf surfaces. Environ. Microbiol. Rep. 4, 209-216.
• Feingersch, R., Philosof, A., Mejuch, T., Glaser, F., Alalouf, O., Shoham, Y., Béjà, O. (2012) Potential for phosphite and phosphonate utilization by Prochlorococcus. ISME J. 6, 827-834.
• Enav, H., Béjà, O., Mandel-Gutfreund, Y. (2012) Cyanophage tRNAs may play a role in cross-infectivity of oceanic Prochlorococcus and Synechococcus hosts. ISME J. 6, 619-628.
• Atamna-Ismaeel, N., Finkel, O.M., Glaser, F., Sharon, I., Schneider, R., Post, A.F., Spudich, J.L., von Mering, C., Vorholt, J.A., Iluz, D., Béjà, O., Belkin, S. (2012) Microbial rhodopsins on leaf surfaces of terrestrial plants. Environ. Microbiol. 14, 140-146.
2011
• Philosof, A., Battchikova, N., Aro, E.-M., Béjà, O. (2011) Marine cyanophages: Tinkering with the electron transport chain. ISME J. 5, 1568-1570.
• Sharon, I., Battchikova, N., Aro, E.-M., Giglione, C., Meinnel, T., Glaser, F., Pinter, R.Y., Breitbart, M., Rohwer, F., Béjà, O. (2011) Comparative metagenomics of microbial traits within oceanic viral communities. ISME J. 5, 1178-1190.
• Yogev, T., Rahav, E., Bar Zeev, E., Aharonovich, D., Stambler, N., Kress, N., Béjà, O., Mulholland, M., Herut, B., Berman-Frank, I. (2011) Is dinitrogen fixation significant in the Levantine Basin, East Mediterranean Sea? Environ. Microbiol. 13, 854-871.
• Alperovitch, A., Sharon, I., Rohwer, F., Aro, E.M., Milo, R., Nelson, N., Béjà, O. (2011) Reconstructing a puzzle: Existence of cyanophages containing both photosystem-I & photosystem-II gene-suites inferred from oceanic metagenomic datasets. Environ. Microbiol. 13, 24-32.
2010
• Koh, Y.E., Atamna-Ismaeel, N., Andrew R. Martin, A.R., Cowie, R.O.M., Béjà, O. Davy, S.K., Ryan, K.G. (2010) Proteorhodopsin-bearing bacteria in Antarctic sea ice. Appl. Environ. Microbiol. 76, 5918-5925.
• DeLong, E.F., and Béjà, O. (2010) The light-driven proton pump proteorhodopsin enhances bacterial survival during tough times. PLoS Biol. 8, e1000359.
• Man-Aharonovich, D., Philosof, A., Kirkup, B.C., Le Gall, F., Yogev, T., Berman-Frank, I., Polz, M.F., Vaulot, D., Béjà, O. (2010) Diversity of active marine picoeukaryotes in the Eastern Mediterranean Sea unveiled using photosystem-II psbA transcripts. ISME J. 4, 1044-1052.
• Bodaker, I., Sharon, I., Suzuki, M.T., Feingersch, R., Shmoish, M., Andreishcheva, E., Sogin, M.L., Rosenberg, M., Maguire, M.E., Belkin, S., Oren, O., Béjà, O. (2010) Comparative community genomics in the Dead Sea, an increasingly extreme environment. ISME J. 4 , 399-407.
• Feingersch, R., Suzuki, M.T., Shmoish, M., Sharon, I., Sabehi, G., Partensky, F., Béjà, O. (2010) Microbial community genomics in eastern Mediterranean Sea surface waters. ISME J. 4, 78-87.
• Mano, A., Tuller, T., Béjà, O., Pinter, R.Y. (2010) Comparative classification of species and the study of pathway evolution based on the alignment of metabolic pathways. BMC Bioinformatics 11, S38.
2009
• Bodaker, I., Béjà, O., Sharon, I., Feingersch, R., Rosenberg, M., Oren, A., Hindiyeh, M.Y., Malkawi, H.I. (2009) Archaeal diversity in the Dead Sea: microbial survival under increasingly harsh conditions. Natural Resources and Environmental Issues V5, Article 25.
• Philosof, A., Sabehi, G., Béjà, O. (2009) Comparative analysis of actinobacterial genomic fragments from Lake Kinneret. Environ. Microbiol. 11, 3189-3200.

• Sharon, I., Alperovitch, A., Rohwer, F., Haynes, M., Glaser, F., Atamna-Ismaeel, N., Pinter, R.Y., Partensky, F., Koonin, E.V., Wolf, Y.I., Nelson, N., Béjà, O. (2009) Photosystem-I gene cassettes are present in marine virus genomes. Nature 461, 258-262.
• Yutin, N., Suzuki, M.T., Rosenberg, M., Rotem, D., Madigan, M.T., Suling, J., Imhoff, J.F., Béjà, O. (2009) BchY-based degenerate primers target all types of anoxygenic photosynthetic bacteria in a single PCR reaction. Appl. Environ. Microbiol. 75, 7556-7559.
• Feingersch, R., and Béjà, O. (2009) Bias in assessments of marine SAR11 biodiversity in environmental fosmid & BAC libraries? ISME J. 3, 1117-1119.
• Tzahor, S., Man-Aharonovich, D., Kirkup, B.C., Yogev, T., Berman-Frank, I., Polz, M., Béjà, O., Mandel-Gutfreund, Y. (2009) A supervised learning approach for taxonomic classification of core-photosystem-II genes and transcripts in the marine environment. BMC Genomics 10,229.
• Loy, A., Duller, S., Baranyi, C., Musmann, M., Ott, J., Sharon, I., Béjà, O., Le Paslier, D., Dahl, C., Wagner, M. (2009) Reverse dissimilatory sulfite reductase and other Dsr Proteins in sulfur-oxidizing bacteria: evolutionary history and suitability as phylogenetic markers. Environ. Microbiol. 11, 289-299.
2008
• Yutin, N., Béjà, O., Suzuki, M.T. (2008) The use of denaturing gradient gel electrophoresis with fully degenerate pufM primers to monitor aerobic anoxygenic phototrophic assemblages. Limnol. Oceanogr. Methods 6, 427-440.
• Bar Zeev, E., Yogev, T., Man-Aharonovich, D., Kress, N., Herut, B., Béjà, O., Berman-Frank, I. (2008) Seasonal dynamics of the endosymbiotic, nitrogen-fixing cyanobacterium Richelia intracellularis in the eastern Mediterranean Sea. ISME J. 2, 911-923.
• Atamna-Ismaeel, N., Sabehi, G., Sharon, I., Witzel, K-P., Labrenz, M., Jurgens, K., Barkay, T., Stomp, M., Huisman, J., Béjà, O. (2008) Widespread distribution of proteorhodopsins in freshwater and brackish ecosystems. ISME J. 2, 656-662.
• Kagan, J., Sharon, I., Béjà, O., Kuhn, J. (2008) The tryptophan pathway genes of the Sargasso Sea metagenome: new operon structures and the prevalence of non-operon organization. Genome Biol. 9, R20
• Massana, R., Karniol, B., Pommier, T., Bodaker, I., Béjà, O. (2008) Metagenomic retrieval of a ribosomal DNA repeat array from an uncultured marine alveolate. Environ. Microbiol. 10, 1335-1343.
• Béjà, O., and Suzuki, M.T. (2008) Photoheterotrophic marine prokaryotes. In Microbial Ecology of the Oceans 131-157. Kirchman, D.L. (ed). New York: John Wiley & Sons, Inc.
2007
• Sharon, I., Tzahor, S., Williamson, S., Shmoish, M., Man-Aharonovich, D., Rusch, D.B., Yooseph, S., Zeidner, G., Golden, S.S., Mackey, S.R., Adir, N., Weingart, U., Horn, D., Venter, J.C, Mandel-Gutfreund, Y., Béjà, O. (2007) Viral photosynthetic reaction center genes and transcripts in the marine environment. ISME J. 1, 492-501.
• Man-Aharonovich, D., Kress, N., Bar Zeev, E., Berman-Frank, I., Béjà, O. (2007) Molecular ecology of nifH genes and transcripts in Eastern Mediterranean Sea. Environ. Microbiol. 9, 2354-2363.
• Yutin, N., Suzuki, M.T., Teeling, H., Weber, M., Venter, J.C., Rusch, D., Béjà, O. (2007) Assessing diversity and biogeography of aerobic anoxygenic phototrophic bacteria in surface waters of the Atlantic and Pacific Oceans using the Global Ocean Sampling expedition metagenomes. Environ. Microbiol. 9, 1464-1475.
• Sabehi, G., Kirkup, B.C., Rosenberg, M., Stambler, N., Polz, M.F., Béjà, O. (2007) Adaptation and spectral tuning in divergent marine proteorhodopsins from the eastern Mediterranean and the Sargasso Seas. ISME J. 1, 48-55.
• Suzuki, M.T., and Béjà, O. (2007) An elusive marine photosynthetic bacterium is finally unveiled. Proc. Natl. Acad. Sci. USA 104, 2561-2562.
• Sabehi, G., and Béjà, O. (2007) BAC libraries construction from marine microbial assemblages. Molecular Microbial Ecology Manual 1863-1879.
2005
• Yutin, N., Suzuki, M.T., Béjà, O. (2005) Novel primers reveal a wider diversity among marine aerobic anoxygenic phototrohs. Appl. Environ. Microbiol. 71, 8958-8962.
• Zeidner, G., and Béjà, O. (2005) Community-level analysis of phototrophy: psbA gene diversity. Methods Enzymol. 397, 372-380.
• Yutin, N., and Béjà, O. (2005) Putative novel photosynthetic reaction center organizations in marine aerobic anoxygenic photosynthetic bacteria: insights from environmental genomics & metagenomics. Environ. Microbiol. 7, 2027-2033.

• Sabehi, G., Loy, A., Jung, K.H., Partha, R., Spudich, J.L., Isaacson, T., Hirschberg, J., Wagner, M., Béjà, O. (2005) New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol. 3, e173.

• Zeidner, G., Bielawski, J.P., Shmoish, M., Scanlan, D.J., Sabehi, G., Béjà, O. (2005) Potential photosynthesis gene recombination between Prochlorococcus and Synechococcus via viral intermediates. Environ. Microbiol. 7, 1505-1513.
• Oz, A., Sabehi, G., Koblizek, M., Massana, R., Béjà, O. (2005) Roseobacter-like bacteria in Red and Mediterranean Sea aerobic anoxygenic photosynthetic populations. Appl. Environ. Microbiol. 71, 344-353.
2004
• Suzuki, M.T., Preston, C.M., Béjà, O., de la Torre, R.J., Steward, G., DeLong, E.F. (2004) Quantitative phylogenetic screening of ribosomal RNA gene-containing clones in Bacterial Artificial Chromosome (BAC) libraries from different depths in Monterey Bay. Microbial Ecol. 48, 473-488.
• Bielawski, J. P., Dunn, K.A., Sabehi, G., Béjà, O. (2004) Darwinian adaptation of proteorhodopsin to different light intensities in the environment. Proc. Natl. Acad. Sci. USA 141, 14824-14829.
• Sabehi, G., Béjà, O., Preston, C. M., Suzuki, M. T., DeLong, E.F. (2004) Different SAR86 subgroups harbour divergent proteorhodopsins. Environ. Microbiol. 6, 903-910.
• Béjà, O. (2004) To BAC or not to BAC: Marine Ecogenomics. Curr. Opin. Biotech. 15, 187-190.
• Man-Aharonovich, D., Sabehi, G., Sineshchekov O.A., Spudich, E.N., Spudich, J.L., Béjà, O. (2004) Characterization of RS29, a blue-green proteorhodopsin variant from the Red Sea. Photochem. Photobiol. Sci. 3, 459-462.
• Zeidner, G., and Béjà, O. (2004) The use of DGGE analyses to explore eastern Mediterranean and Red Sea marine picophytoplankton assemblages. Environ. Microbiol. 6, 528-534.
2003
• Koblizek M., Béjà, O., Bidigare R.R., Christensen S., Benitez-Nelson B., Vetriani C., Kolber M.K., Falkowski P.G., Kolber Z.S. (2003) Isolation and Characterization of Erythrobacter sp. Strains from the Upper Ocean. Arch. Microbiol. 180, 327-338.
• de la Torre, R.J., Christianson, L., Béjà, O., Suzuki, M.T., Karl, D., Heidelberg, J.F., DeLong, E.F. (2003) Proteorhodopsin genes are widely distributed among divergent bacterial taxa. Proc. Natl. Acad. Sci. USA 100, 12830-12835.
• Sabehi, G., Massana, R., Bielawski J.P., Rosenberg M., Delong, E.F., Béjà, O. (2003) Novel Proteorhodopsin Variants from the Mediterranean and Red Seas. Environ. Microbiol. 5, 842-849.

• Man, D., Wang, W., Sabehi, G., Aravind, L., Post, A.F., Massana, R., Spudich, E.N., Spudich, J.L., Béjà, O. (2003) Diversification and spectral tuning in marine proteorhodopsins. EMBO J. 22, 1725-1731.
• Zeidner, G., Preston, C.M., DeLong, E.F., Massana, R., Post, A.F., Scanlan, D.J., Béjà, O. (2003) Molecular diversity among marine picophytoplankton as revealed by psbA analyses. Environ. Microbiol. 5, 212-216.
2002

• Béjà, O., Suzuki, M.T., Heidelberg, J.F., Nelson, W.C., Preston, C.M., Hamada, T., Eisen, J.A., Fraser, C.M., DeLong, E.F. (2002). Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415, 630-633.
• Béjà, O., Koonin, E.V., Aravind, L., Taylor, L.T., Seitz, H., Stein, J.L., Bensen, D.C., Feldman, R.A., Swanson, R.V., DeLong, E.F. (2002). Comparative genomic analysis of archaeal genotypic variants in a single population, and in two different oceanic provinces. Appl. Environ. Microbiol. 68, 335-345.
2001

• Béjà*, O., Spudich*, E.N., Spudich, J.L., Leclerc, M., DeLong, E.F. (2001). Proteorhodopsin phototrophy in the ocean. Nature 411, 786-789.
• Suzuki, M.T., Béjà, O., Taylor, L.T., DeLong, E.F. (2001). Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environ. Microbiol. 3, 323-331.
2000

• Béjà, O., Aravind, L., Koonin, E.V., Suzuki, M.T., Hadd, A., Nguyen, C., Jovanovich, S. B., Gates, C.M., Feldman, R.A., Spudich, J.L., Spudich, E.N., DeLong, E.F. (2000). Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289, 1902-1906.
• Béjà, O., Suzuki, M.T., Koonin, E.V., Aravind, L., Hadd, A., Nguyen, L.P., Villacorta, R., Amjadi, M., Garrigues, C., Jovanovich, S.B., Feldman, R.A., DeLong, E.F. (2000). Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2, 516-529.

