coming soon
• Takaramoto, S., Fainsod, S., Nagata, T., Rozenberg, A., Béjà, O., Inoue, K. Significance of extended N-terminal region and a first motif residue in a third transmembrane helix of a novel-cation channelrhodopsin, HulaCCR. in preparation
• Chazan, A., et al. Viral-encoded UV-sensing heliorhodopsin proteins XXXX. in preparation
• Yaish, S., et al. Suggested roles for heliorhodopsin in Lactococcus lactis strain 21. in preparation
• Tzlil, G., et al. Light-harvesting by antenna-containing rhodopsins in uncultured marine XXXX. in preparation
• Nadel, O., Hanna, R., Rozenberg, A., Suissa-Szlejf, M., Sulimani, L., Adir, N., Béjà, O., Kleifeld, O. An abundant uncultured marine cyanophage family encodes two distinct NblA proteins with differential activities toward different cyanobacterial phycobiliprotein subunits. in preparation
• Nadel, O., Hanna, R., Rozenberg, A., Shitrit, D., Tahan, R., Pakarsky, I., Kleifeld, O., Lindell, D., Béjà, O. Oceanic photosynthesis is directly affected by viral NblA mediated degradation of cyanobacterial phycobilisomes. in preparation
2024
• Wegner, H., Roitman, S., Kupczok, A., Braun, V., Woodhouse, J., Grossart, H-.P., Zehner, S., Béjà, O., and Frankenberg-Dinkel, N. Viral encoded Shemin pathway points towards the importance of tetrapyrrole metabolism during infection. Nat. Commun. in revision
see BioRxiv deposition
2023
• Mannen, K., Nagata, T., Rozenberg, A., Konno, M., Marín, M.d.C., Bagherzadeh, R., Béjà, O., Uchihashi, T., Inoue, K. (2023) Multiple roles of a conserved glutamate residue for unique biophysical properties in a new group of microbial rhodopsins homologous to TAT rhodopsin. J. Mol. Biol. 436, 168331
• Oppermann, J., Rozenberg, A., Fabrin, T., Gonzalez Cabrera, C., Béjà, O., Prigge, M., Hegemann, P. (2023) Robust Optogenetic inhibition with red-light-sensitive anion-conducting channelrhodopsins. eLife 10.7554/eLife.90100.1
• News & Views by Béjà, O., and Inoue, K. (2023) Iron-limitation light switch. Nat. Microbiol. 8, 1942-1943
• Bar-Shalom*, R., Rozenberg*, A., Lahyani, M., Hassanzadeh, B., Sahoo, G., Haber, M., Burgsdorf, I., Tang, X., Squattrito, V., Gomez-Consarnau, L., Béjà, O., Steindler, L. (2023) Rhodopsin-mediated nutrient uptake by cultivated photoheterotrophic Verrucomicrobiota. ISME J. 17, 1063-1073
*equal contribution

• Chazan*, A., Das*, I., Fujiwara*, T., Shunya*, M., Rozenberg*, A., Molina-Márquez, A., Fumiya, K.S., Tanaka, T., Gómez-Villegas, P., Larom, S., Pushkarev, A., Malakar, P., Hasegawa, M., Tsukamoto, Y., Ishizuka, T., Konno, M., Nagata, T., Mizuno, Y., Katayama, K., Abe-Yoshizumi, R., Ruhman, S., Inoue, K., Kandori, H., León, R., Shihoya, W., Yoshizawa, S., Sheves, M., Nureki, O., Béjà, O. (2023) Phototrophy by antenna-containing rhodopsin pumps in aquatic environments. Nature 615, 535-540
*equal contribution
see also our BEHIND THE PAPER story.
• Roitman, S., Rozenberg, A., Lavy, T., Brussaard, C.P.D., Kleifeld, O., and Béjà, O. (2023) Isolation and infection cycle of a polinton-like virus virophage in an abundant marine alga. Nat. Microbiol. 8, 332-346
see also our BEHIND THE PAPER story.
2022
• Vierock, J., Peter, E., Grimm, C., Rozenberg, A., Chen, I.W., Tillert, L., Castro Scalise, A.G., Casini, M., Augustin, S., Tanese, D., Forget, B.C., Peyronnet, R., Schnider-Warme, F., Emiliani, V., Béjà, O., Hegemann, P. (2022) WiChR, a highly potassium selective channelrhodopsin for low-light one- and two-photon inhibition of excitable cells. Sci. Adv. 8, eadd7729
• Hososhima, S., Mizutori, R., Abe-Yoshizumi, R., Rozenberg, A., Shigemura, S., Pushkarev, A., Konno, M., Katayama, K., Inoue, K., Tsunoda, S.P., Béjà, O., and Kandori, H. (2022) Proton-transporting heliorhodopsins from marine giant viruses. eLife 11, e78416
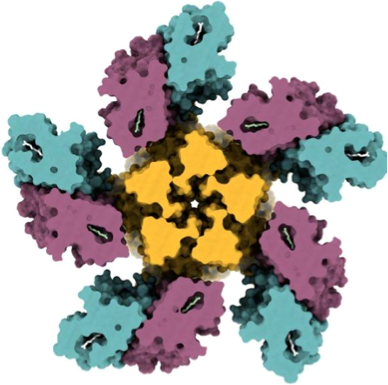
• Rozenberg*, A., Kaczmarczyk*, I., Matzov*, D., Vierock*, J., Nagata, T., Sugiura, M., Katayama, K., Kawasaki, Y., Konno, M., Nagasaki, Y., Toyama, M., Das, I., Pahima, E., Church, J., Adam, S., Borin, V.A., Chazan, A., Augustin, S., Wietek, J., Dine, J., Peleg, Y., Kawanabe, A., Fujiwara, Y., Yizhar, O., Sheves, M., Shapiro, I., Furutani, Y., Kandori, H., Inoue, K., Hegemann, P., Béjà, O., and Shalev-Benami, M. (2022) Rhodopsin-bestrophin fusion proteins from unicellular algae form gigantic pentameric ion channels. Nat. Struct. Mol. Biol. 29, 592-603.
*equal contribution
• Rodriguez-Valera, F., Pushkarev, A., Rosselli, R., and Béjà, O. (2022) Searching metagenoms for new rhodopsins. In: Gordeliy, V. (ed.) Rhodopsin. Methods Molecular Biology, 2501, 101-108.
• Jaffe, A.L., Konno, M., Kawasaki, Y., Kataoka, C., Béjà, O., Kandori, H., Inoue, K., and Banfield, J.F. (2022) Saccharibacteria harness light energy using Type-1 rhodopsins that may rely on retinal sourced from microbial hosts. ISME J. 16, 2056-2059.
• Chazan*, A., Rozenberg*, A., Mannen, K., Nagata, T., Tahan, R., Yaish, S., Larom, S., Inoue, K., Béjà, O., and Pushkarev, A. (2022) Diverse heliorhodopsins detected via functional metagenomics in freshwater Actinobacteria, Chloroflexi and Archaea. Environ. Microbiol. 24, 110-121.
*equal contribution
2021
• Rozenberg*, A., Inoue*, K., Kandori, H., and Béjà, O. (2021) Microbial rhodopsins: the last two decades. Annu. Rev. Microbiol. 75, 427-447
*equal contribution
• Dispatch by Arava, Y., and Béjà, O. (2021) Phage biology: Stuck with dU. Curr. Biol. 31, R898-R899.
• Inoue, K., Karasuyama, M., Nakamura, R., Konno, M., Yamada, D., Mannen, K., Nagata, T., Inatsu, Y., Yawo, H., Yura, K., Béjà, O., Kandori, H., and Takeuchi, I. (2021) Exploration of natural red-shifted rhodopsins by a machine-learning based Bayesian experimental design. Commun. Biol. 4, 362.
• Kataoka, C., Sugimoto, T., Shigemura, S., Katayama, K., Tsunoda, S.P., Inoue, K., Béjà, O., and Kandori, H. (2021) TAT rhodopsin is a UV-dependent environmental pH sensor. Biochemistry. 60, 899-907.
2020
•Hevroni, G., Flores-Uribe, J., Béjà, O., and Philosof, A. (2020) Seasonal and diel patterns of abundance and activity of viruses in the Red Sea. Proc. Natl. Acad. Sci. USA. .
 see also our BEHIND THE PAPER story.
see also our BEHIND THE PAPER story.
• Rozenberg, A., Oppermann J., Wietek, J., Fernandez Lahore, R.G., Sandaa, R.-A., Bratbak, G., Hegemann, P., and Béjà, O. (2020) Lateral gene transfer of anion-conducting channelrhodopsins between green algae and giant viruses. Curr. Biol. 30, 4910-4920.
 see also our BEHIND THE PAPER story.
see also our BEHIND THE PAPER story.
see Dispatch by Gallot-Lavallée, L., and Archibald, J.M. (2020) Evolution: Viral rhodopsins illuminate algal evolution.
• Inoue, K., Tsunoda, S.P., Singh, M., Tomida, S., Hososhima, S., Konno, M., Nakamura, R., Watanabe, H., Uchihashi, T., Bulzu, P.-A., Banciu, H.L., Andrei, A.-Ş., Ghai, R., Béjà, O., and Kandori, H. (2020) Schizorhodopsins: A family of rhodopsins from Asgard archaea that function as light-driven inward H+pumps. Sci. Adv. 6, eaaz2441.
• Pan, J., Zhou, Z., Béjà, O., Cai, M., Yang, Y., Liu, Y., Gu, J.-D., and Li, M. (2020) Genetic and transcriptomic evidence of light sensing, porphyrin biosynthesis, Calvin-Benson-Bassham cycle, and urea production in Bathyarchaeota. Microbiome 8, 43.
2019
• Nadel, O., Rozenberg, A., Flores-Uribe, J., Larom, S., Schwarz, R., and Béjà, O. (2019) An uncultured marine cyanophage encodes an active phycobilisome proteolysis adaptor protein NblA. Environ. Microbiol. Rep. 11, 848-854.
• Singh, M., Katayama, K. Béjà, O., and Kandori, H. (2019) Anion binding to mutants of the Schiff base counterion in heliorhodopsin 48C12. Phys. Chem. Chem. Phys. 21, 23663.

• Shihoya, W., Inoue, K., Singh, M., Konno, M., Hososhima, S., Yamashita, K., Ikeda, K., Higuchi, A., Izume, T., Okazaki, S., Hashimoto, M., Mizutori, R., Tomida, S., Yamaguchi, Y., Abe-Yoshizumi, R., Katayama, K., Tsunoda, S.P., Shibata, M., Furutani, Y., Pushkarev, A., Béjà, O., Uchihashi, T., Kandori, H., and Nureki, O. (2019) Crystal structure of heliorhodopsin. Nature 574, 132-136.
• Karaoke, C., Inoue, K., Katayama, K., Béjà, O., and Kandori, H. (2019) Unique photochemistry observed in a new microbial rhodopsin. J. Phys. Chem. Lets. 10, 5117-5121.
• Oppermann, J., Liepe, B., Fischer, P., Rodriguez-Rozada, S., Flores-Uribe, J., Salipetre, A., Peter, E., Keidel, A., Vierock, J., Kaufmann, J., Broser, M., Luck, M., Bartl, F., Hildebrandt, P., Wiegert, J.S., Béjà, O., Hegemann, P., and Wietek, J. (2019) MerMAIDs: A novel family of metagenomically discovered, marine, anion-conducting and intensely desensitizing channelrhodopsins. Nature Commun. 10, 3315.
 see also our BEHIND THE PAPER story.
see also our BEHIND THE PAPER story.
• Yamaguchi, Y., Konno, M., Yamada, D., Yura, K., Inoue, K., Béjà, O., and Kandori, H. (2019) Engineered functional recovery of microbial rhodopsin without retinal-binding lysine. Photochem. Photobiol. 95, 116-1121.
• Flores-Uribe, J., Philosof, A., Sharon, I., Fridman, S., Larom, S., and Béjà, O. (2019) A novel uncultured marine cyanophage lineage with lysogenic potential linked to a putative marine Synechococcus ‘relic’ prophage. Environ. Microbiol. Rep. 11, 598-604.
• Bulzu, P.-A., Andrei, A.-Ş., Salcher, M.M., Mehrshad, M., Inoue, K., Kandori, H., Béjà, O., Ghai, R., and Banciu, H.L. (2019) Casting light on Asgardarchaeota metabolism in a sunlit microoxic niche. Nature Microbiol. 4, 1129-1137.
 see also our BEHIND THE PAPER story.
see also our BEHIND THE PAPER story.
• Tahara, S., Singh, M., Kuramochi, H., Shihoya, W., Inoue, K., Nureki, O., Béjà, O., Mizutani, Y., Kandori, H., and Tahara, T. (2019) Ultrafast dynamics of heliorhodopsins. J. Phys. Chem. B 123, 2507-2512.
• Flores-Uribe*, J., Hevroni*, G., Ghai, R., Pushkarev, A., Inoue, K., Kandori, H.,and Béjà, O. (2019) Heliorhodopsins are absent in diderm (Gram-negative) bacteria: Some thoughts and possible implications for activity. Environ. Microbiol. Rep. 11, 419-424.
*equal contribution
 see also our BEHIND THE PAPER story.
see also our BEHIND THE PAPER story.
2018

• Enav, H., Kirzner, S., Lindell, D., Mandel-Gutfreund, Y., and Béjà, O. (2018) Adaptation to sub-optimal hosts is a driver of viral diversification in the ocean. Nature Commun. 9, 4698.
• Otomo, A., Mizuno, M., Singh, M., Shihoya, W., Inoue, K., Nureki, O., Béjà, O., Kandori, H., Mizutani, Y. (2018) Resonance Raman investigation of the chromophore structure of heliorhodopsins. J. Phys. Chem. Lett. 8, 6431-6436.
• Singh, S., Inoue, K., Pushkarev, A., Béjà, O., and Kandori, H. (2018) Mutation study of heliorhodopsin 48C12. Biochemistry 57, 5041-5049.

• Pushkarev, A., Inoue, K., Larom, S., Flores-Uribe, J., Singh, M., Konno, M., Tomida, S., Ito, S., Nakamura, R., Tsunoda, S.P., Philosof, A., Sharon, I., Yutin, N., Koonin, E.V., Kandori K., and Béjà, O. (2018) A distinct abundant group of microbial rhodopsins discovered using functional metagenomics. Nature 558, 595-599.
 see also our BEHIND THE PAPER story.
see also our BEHIND THE PAPER story.
• Pushkarev, A., Hevroni, G., Roitman, S., Shim, J.-G., Choi, A., Jung, K.-H., and Béjà, O. (2018) The use of a chimeric rhodopsin vector for the detection of new proteorhodopsins based on color. Front. Microbiol. 9, 439.
• Ledermann, B., Schwan, M., Sommerkamp, J., Hofmann, E., Béjà, O., and Frankenberg-Dinkel, N. (2018) Evolution and molecular mechanisms of four-electron reducing ferredoxin-dependent bilin reductase from oceanic phages. FEBS J. 285, 339-356.
• Roitman, S., Hornung, E., Flores-Uribe, J., Sharon, I., Feussner, I., and Béjà, O. (2018) Cyanophage-encoded lipid-desaturases: oceanic distribution, diversity and function. ISME J. 12, 343-355.
2017

• Fridman, S., Flores-Uribe, J., Larom, S., Alalouf, O., Liran, O., Yacoby, I., Salama, F., Bailleul, B., Rappaport, F., Ziv, T., Sharon, I., Cornejo-Castillo, F.M., Philosof, A., Dupont, C.L., Sanchez, P., Acinas, S.G., Rohwer, F., Lindell, D. and Béjà, O. (2017) A myovirus encoding both photosystem-I and II proteins enhances cyclic electron flow in infected Prochlorococcus cells. Nat. Microbiol. 2, 1350-1357.
 see also BEHIND THE PAPER.
see also BEHIND THE PAPER.
• Philosof, A., Yutin, N., Flores-Uribe, J., Sharon, I., Koonin, E.V. and Béjà, O. (2017) Novel abundant oceanic viruses of uncultured Marine Group II Euryarchaeota. Curr. Biol. 27, 1362-1368.
• Yaniv, K., Golberg, K., Kramarsky-Winter, E., Marks, R., Pushkarev, A., Béjà, O., and Kushmaro, A. (2017) Functional marine metagenomic screening for anti-quorum sensing and anti-biofilm activity. Biofouling 33, 1-13.
2016
• Pinhassi, J., DeLong, E.F., Béjà, O., Gonzalez, J.M., and Pedros-Alio, C. (2016) Marine bacterial and archaeal ion-pumping rhodopsins: genetic diversity, physiology and ecology. Microbiol. & Mol. Biol. Rev. 80, 929-954. ![]()
• Ledermann, B., Béjà, O., and Frankenberg-Dinkel, N. (2016) New biosynthetic pathway for pink pigments from uncultured oceanic viruses. Environ. Microbiol. 18, 4337-4347. ![]()
• Pushkarev, A., and Béjà, O. (2016) Functional metagenomic screen reveals new and diverse microbial rhodopsins. ISME J. 10, 2331-2335. ![]()
2015
• Roitman*, S., Flores-Uribe*, J., Philosof, A., Knowles, B., Rohwer, F., Ignacio-Espinoza, J.C., Sullivan, M.B., Cornejo-Castillo, F.M., Sanchez, P., Acinas, S.G., Dupont, C.L., and Béjà, O. (2015) Closing the gaps on the viral photosystem-I psaDCAB gene organization. Environ. Microbiol. 17, 5100-5108. ![]()
*equal contribution
• Hevroni*, G., Enav*, H., Rohwer, F., and Béjà, O. (2015) Diversity of viral photosystem-I psaA genes. ISME J. 9, 1892-1898.
*equal contribution
2014
• Beja O. and Lanyi, J.K. (2014) Nature’s toolkit for microbial rhodopsin ion pumps. Proc. Natl. Acad. Sci. USA 111, 6538-6539. ![]()
• Enav, H., Mandel-Gutfreund, Y., and Béjà, O. (2014) Comparative metagenomic analyses reveal viral induced shifts of host metabolism towards nucleotide biosynthesis. Microbiome 2, 9.
2013
• Sabehi, G., and Béjà, O. (2013) Preparation of BAC libraries from marine microbial populations. Methods Enzymol. 531, 111-122.
• Philosof, A., and Béjà, O. (2013) Bacterial, archaeal and viral-like rhodopsins from the Red Sea. Environ. Microbiol. Rep. 5, 475-482. ![]()
• Béjà, O., Pinhassi, J. and Spudich, J.L. Proteorhodopsins: widespread microbial light-driven proton pumps. In: Levin S.A. (ed.) Encyclopedia of Biodiversity 6, 280-285.
• Finkel, O.M., Béjà, O., and Belkin, S. (2013) Global abundance of microbial rhodopsins. ISME J. 7, 448-451.
2012
• Mazor, Y., Greenberg, I., Toporik, H., Béjà, O., and Nelson, N. (2012) The evolution of PSI in light of phage encoded reaction centers. Phil. Trans. R. Soc. B. 367, 3400-3405.
• Bodaker, I., Suzuki, M.T., Oren, O., and Béjà, O. (2012) Dead Sea rhodopsins revisited. Environ. Microbiol. Rep. 4, 617-621.
• Béjà, O., Fridman, S., and Glaser, F. (2012) Viral clones from the GOS expedition with an unusual photosystem-I gene cassette organization. ISME J. 6, 1617-1620.
• Atamna-Ismaeel, N., Finkel, O.M., Glaser, F., von Mering, C., Vorholt, J.A., Koblizek, M., Belkin, S., and Béjà, O. (2012) Bacterial anoxygenic photosynthesis on plant leaf surfaces. Environ. Microbiol. Rep. 4, 209-216.
• Feingersch, R., Philosof, A., Mejuch, T., Glaser, F., Alalouf, O., Shoham, Y. and Béjà, O. (2012) Potential for phosphite and phosphonate utilization by Prochlorococcus. ISME J. 6, 827-834.
• Enav, H., Béjà, O., and Mandel-Gutfreund, Y. (2012) Cyanophage tRNAs may play a role in cross-infectivity of oceanic Prochlorococcus and Synechococcus hosts. ISME J. 6, 619-628.
• Atamna-Ismaeel, N., Finkel, O.M., Glaser, F., Sharon, I., Schneider, R., Post, A.F., Spudich, J.L., von Mering, C., Vorholt, J.A., Iluz, D., Béjà, O. and Belkin, S. (2012) Microbial rhodopsins on leaf surfaces of terrestrial plants. Environ. Microbiol. 14, 140-146.
2011
• Philosof, A., Battchikova, N., Aro, E.-M., and Béjà, O. (2011) Marine cyanophages: Tinkering with the electron transport chain. ISME J. 5, 1568-1570.
• Sharon, I., Battchikova, N., Aro, E.-M., Giglione, C., Meinnel, T., Glaser, F., Pinter, R.Y., Breitbart, M., Rohwer, F., and Béjà, O. (2011) Comparative metagenomics of microbial traits within oceanic viral communities. ISME J. 5, 1178-1190.
• Yogev, T., Rahav, E., Bar Zeev, E., Aharonovich, D., Stambler, N., Kress, N., Béjà, O., Mulholland, M., Herut, B., and Berman-Frank, I. (2011) Is dinitrogen fixation significant in the Levantine Basin, East Mediterranean Sea? Environ. Microbiol. 13, 854-871.
• Alperovitch*, A., Sharon*, I., Rohwer, F., Aro, E.M., Milo, R., Nelson, N., and Béjà, O. (2011) Reconstructing a puzzle: Existence of cyanophages containing both photosystem-I & photosystem-II gene-suites inferred from oceanic metagenomic datasets. Environ. Microbiol. 13, 24-32.
*equal contribution
2010
• Koh, Y.E., Atamna-Ismaeel, N., Andrew R. Martin, A.R., Cowie, R.O.M., Béjà, O. Davy, S.K., and Ryan, K.G. (2010) Proteorhodopsin-bearing bacteria in Antarctic sea ice. Appl. Environ. Microbiol. 76, 5918-5925.
• DeLong, E.F., and Béjà, O. (2010) The light-driven proton pump proteorhodopsin enhances bacterial survival during tough times. PLoS Biol. 8, e1000359.
• Man-Aharonovich*, D., Philosof*, A., Kirkup, B.C., Le Gall, F., Yogev, T., Berman-Frank, I., Polz, M.F., Vaulot, D., and Béjà, O. (2010) Diversity of active marine picoeukaryotes in the Eastern Mediterranean Sea unveiled using photosystem-II psbA transcripts. ISME J. 4, 1044-1052.
*equal contribution
• Bodaker*, I., Sharon*, I., Suzuki, M.T., Feingersch, R., Shmoish, M., Andreishcheva, E., Sogin, M.L., Rosenberg, M., Maguire, M.E., Belkin, S., Oren, O., and Béjà, O. (2010) Comparative community genomics in the Dead Sea, an increasingly extreme environment. ISME J. 4 , 399-407.
*equal contribution
• Feingersch, R., Suzuki, M.T., Shmoish, M., Sharon, I., Sabehi, G., Partensky, F., and Béjà, O. (2010) Microbial community genomics in eastern Mediterranean Sea surface waters. ISME J. 4, 78-87.
• Mano, A., Tuller, T., Béjà, O., and Pinter, R.Y. (2010) Comparative classification of species and the study of pathway evolution based on the alignment of metabolic pathways. BMC Bioinformatics 11, S38.
2009
• Bodaker, I., Béjà, O., Sharon, I., Feingersch, R., Rosenberg, M., Oren, A., Hindiyeh, M.Y., and Malkawi, H.I. (2009) Archaeal diversity in the Dead Sea: microbial survival under increasingly harsh conditions. Natural Resources and Environmental Issues V5, Article 25.
• Philosof, A., Sabehi, G., and Béjà, O. (2009) Comparative analysis of actinobacterial genomic fragments from Lake Kinneret. Environ. Microbiol. 11, 3189-3200.

• Sharon*, I., Alperovitch*, A., Rohwer, F., Haynes, M., Glaser, F., Atamna-Ismaeel, N., Pinter, R.Y., Partensky, F., Koonin, E.V., Wolf, Y.I., Nelson, N. and Béjà, O. (2009) Photosystem-I gene cassettes are present in marine virus genomes. Nature 461, 258-262.
*equal contribution
• Yutin, N., Suzuki, M.T., Rosenberg, M., Rotem, D., Madigan, M.T., Suling, J., Imhoff, J.F., and Béjà, O. (2009) BchY-based degenerate primers target all types of anoxygenic photosynthetic bacteria in a single PCR reaction. Appl. Environ. Microbiol. 75, 7556-7559.
• Feingersch, R., and Béjà, O. (2009) Bias in assessments of marine SAR11 biodiversity in environmental fosmid & BAC libraries? ISME J. 3, 1117-1119.
• Tzahor*, S., Man-Aharonovich*, D., Kirkup, B.C., Yogev, T., Berman-Frank, I., Polz, M., Béjà, O., and Mandel-Gutfreund, Y. (2009) A supervised learning approach for taxonomic classification of core-photosystem-II genes and transcripts in the marine environment. BMC Genomics 10,229.
*equal contribution
• Loy, A., Duller, S., Baranyi, C., Musmann, M., Ott, J., Sharon, I., Béjà, O., Le Paslier, D., Dahl, C., and Wagner, M. (2009) Reverse dissimilatory sulfite reductase and other Dsr Proteins in sulfur-oxidizing bacteria: evolutionary history and suitability as phylogenetic markers. Environ. Microbiol. 11, 289-299.
2008
• Yutin, N., Béjà, O., and Suzuki, M.T. (2008) The use of denaturing gradient gel electrophoresis with fully degenerate pufM primers to monitor aerobic anoxygenic phototrophic assemblages. Limnol. Oceanogr. Methods 6, 427-440.
• Bar Zeev, E., Yogev, T., Man-Aharonovich, D., Kress, N., Herut, B., Béjà, O. and Berman-Frank, I. (2008) Seasonal dynamics of the endosymbiotic, nitrogen-fixing cyanobacterium Richelia intracellularis in the eastern Mediterranean Sea. ISME J. 2, 911-923.
• Atamna-Ismaeel, N., Sabehi, G., Sharon, I., Witzel, K-P., Labrenz, M., Jurgens, K., Barkay, T., Stomp, M., Huisman, J., and Béjà, O. (2008) Widespread distribution of proteorhodopsins in freshwater and brackish ecosystems. ISME J. 2, 656-662.
• Kagan, J., Sharon, I., Béjà, O., and Kuhn, J. (2008) The tryptophan pathway genes of the Sargasso Sea metagenome: new operon structures and the prevalence of non-operon organization. Genome Biol. 9, R20
• Massana, R., Karniol, B., Pommier, T., Bodaker, I., and Béjà, O. (2008) Metagenomic retrieval of a ribosomal DNA repeat array from an uncultured marine alveolate. Environ. Microbiol. 10, 1335-1343.
• Béjà, O., and Suzuki, M.T. (2008) Photoheterotrophic marine prokaryotes. In Microbial Ecology of the Oceans 131-157. Kirchman, D.L. (ed). New York: John Wiley & Sons, Inc.
2007
• Sharon*, I., Tzahor*, S., Williamson*, S., Shmoish, M., Man-Aharonovich, D., Rusch, D.B., Yooseph, S., Zeidner, G., Golden, S.S., Mackey, S.R., Adir, N., Weingart, U., Horn, D., Venter, J.C, Mandel-Gutfreund, Y., and Béjà, O. (2007) Viral photosynthetic reaction center genes and transcripts in the marine environment. ISME J. 1, 492-501.
*equal contribution
• Man-Aharonovich, D., Kress, N., Bar Zeev, E., Berman-Frank, I., and Béjà, O. (2007) Molecular ecology of nifH genes and transcripts in Eastern Mediterranean Sea. Environ. Microbiol. 9, 2354-2363.
• Yutin, N., Suzuki, M.T., Teeling, H., Weber, M., Venter, J.C., Rusch, D. and Béjà, O. (2007) Assessing diversity and biogeography of aerobic anoxygenic phototrophic bacteria in surface waters of the Atlantic and Pacific Oceans using the Global Ocean Sampling expedition metagenomes. Environ. Microbiol. 9, 1464-1475.
• Sabehi*, G., Kirkup*, B.C., Rosenberg, M., Stambler, N., Polz, M.F., and Béjà, O. (2007) Adaptation and spectral tuning in divergent marine proteorhodopsins from the eastern Mediterranean and the Sargasso Seas. ISME J. 1, 48-55.
*equal contribution
• Suzuki, M.T., and Béjà, O. (2007) An elusive marine photosynthetic bacterium is finally unveiled. Proc. Natl. Acad. Sci. USA 104, 2561-2562.
• Sabehi, G., and Béjà, O. (2007) BAC libraries construction from marine microbial assemblages. Molecular Microbial Ecology Manual 1863-1879.
2005
• Yutin, N., Suzuki, M.T., and Béjà, O. (2005) Novel primers reveal a wider diversity among marine aerobic anoxygenic phototrohs. Appl. Environ. Microbiol. 71, 8958-8962.
• Zeidner, G., and Béjà, O. (2005) Community-level analysis of phototrophy: psbA gene diversity. Methods Enzymol. 397, 372-380.
• Yutin, N., and Béjà, O. (2005) Putative novel photosynthetic reaction center organizations in marine aerobic anoxygenic photosynthetic bacteria: insights from environmental genomics & metagenomics. Environ. Microbiol. 7, 2027-2033.

• Sabehi*, G., Loy*, A., Jung, K.H., Partha, R., Spudich, J.L., Isaacson, T., Hirschberg, J., Wagner, M., and Béjà, O. (2005) New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol. 3, e173.
*equal contribution

• Zeidner, G., Bielawski, J.P., Shmoish, M., Scanlan, D.J., Sabehi, G., and Béjà, O. (2005) Potential photosynthesis gene recombination between Prochlorococcus & Synechococcus via viral intermediates. Environ. Microbiol. 7, 1505-1513.
• Oz, A., Sabehi, G., Koblizek, M., Massana, R., and Béjà, O. (2005) Roseobacter-like bacteria in Red and Mediterranean Sea aerobic anoxygenic photosynthetic populations. Appl. Environ. Microbiol. 71, 344-353.
2004
• Suzuki, M.T., Preston, C.M., Béjà, O., de la Torre, R.J., Steward, G, and DeLong, E.F. (2004) Quantitative phylogenetic screening of ribosomal RNA gene-containing clones in Bacterial Artificial Chromosome (BAC) libraries from different depths in Monterey Bay. Microbial Ecol. 48, 473-488.
• Bielawski, J. P., Dunn, K.A., Sabehi, G., and Béjà, O. (2004) Darwinian adaptation of proteorhodopsin to different light intensities in the environment. Proc. Natl. Acad. Sci. USA 141, 14824-14829.
• Sabehi, G., Béjà, O., Preston, C. M., Suzuki, M. T., and DeLong, E.F. (2004) Different SAR86 subgroups harbour divergent proteorhodopsins. Environ. Microbiol. 6, 903-910.
• Béjà, O. (2004) To BAC or not to BAC: Marine Ecogenomics. Curr. Opin. Biotech. 15, 187-190.
• Man-Aharonovich, D., Sabehi, G., Sineshchekov O.A., Spudich, E.N., Spudich, J.L., and Béjà, O. (2004) Characterization of RS29, a blue-green proteorhodopsin variant from the Red Sea. Photochem. Photobiol. Sci. 3, 459-462.
• Zeidner, G., and Béjà, O. (2004) The use of DGGE analyses to explore eastern Mediterranean and Red Sea marine picophytoplankton assemblages. Environ. Microbiol. 6, 528-534.
2003
• Koblizek M., Béjà, O., Bidigare R.R., Christensen S., Benitez-Nelson B., Vetriani C., Kolber M.K., Falkowski P.G., Kolber Z.S. (2003) Isolation and Characterization of Erythrobacter sp. Strains from the Upper Ocean. Arch. Microbiol. 180, 327-338.
• de la Torre, R.J., Christianson, L., Béjà, O., Suzuki, M.T., Karl, D., Heidelberg, J.F., and DeLong, E.F. (2003) Proteorhodopsin genes are widely distributed among divergent bacterial taxa. Proc. Natl. Acad. Sci. USA 100, 12830-12835.
• Sabehi, G., Massana, R., Bielawski J.P., Rosenberg M., Delong, E.F., and Béjà, O. (2003) Novel Proteorhodopsin Variants from the Mediterranean and Red Seas. Environ. Microbiol. 5, 842-849.
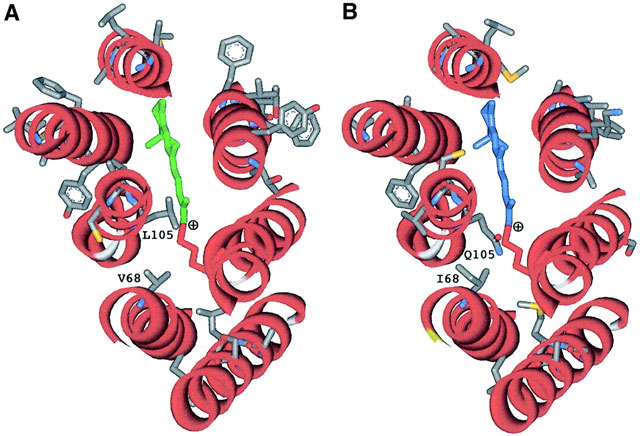
• Man, D., Wang, W., Sabehi, G., Aravind, L., Post, A.F., Massana, R., Spudich, E.N., Spudich, J.L., and Béjà, O. (2003) Diversification and spectral tuning in marine proteorhodopsins. EMBO J. 22, 1725-1731.
• Zeidner, G., Preston, C.M., DeLong, E.F., Massana, R., Post, A.F., Scanlan, D.J., and Béjà, O. (2003) Molecular diversity among marine picophytoplankton as revealed by psbA analyses. Environ. Microbiol. 5, 212-216.
2002

• Béjà, O., Suzuki, M.T., Heidelberg, J.F., Nelson, W.C., Preston, C.M., Hamada, T., Eisen, J.A., Fraser, C.M. and DeLong, E.F. (2002). Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415, 630-633. ![]()
• Béjà, O., Koonin, E.V., Aravind, L., Taylor, L.T., Seitz, H., Stein, J.L., Bensen, D.C., Feldman, R.A., Swanson, R.V. and DeLong, E.F. (2002). Comparative genomic analysis of archaeal genotypic variants in a single population, and in two different oceanic provinces. Appl. Environ. Microbiol. 68, 335-345.
2001

• Béjà*, O., Spudich*, E.N., Spudich, J.L., Leclerc, M., and DeLong, E.F. (2001). Proteorhodopsin phototrophy in the ocean. Nature 411, 786-789. ![]()
*equal contribution
• Suzuki, M.T., Béjà, O., Taylor, L.T., and DeLong, E.F. (2001). Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environ. Microbiol. 3, 323-331.
2000

• Béjà, O., Aravind, L., Koonin, E.V., Suzuki, M.T., Hadd, A., Nguyen, C., Jovanovich, S. B., Gates, C.M., Feldman, R.A., Spudich, J.L., Spudich, E.N., and DeLong, E.F. (2000). Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289, 1902-1906. ![]()
See also Aparna Sreenivasan’s Science Note
• Béjà, O., Suzuki, M.T., Koonin, E.V., Aravind, L., Hadd, A., Nguyen, L.P., Villacorta, R., Amjadi, M., Garrigues, C., Jovanovich, S.B., Feldman, R.A., and DeLong, E.F. (2000). Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2, 516-529.

